We use cutting-edge technologies such as electrophysiology, imaging, and molecular biological techniques to clone novel channels and define the functions of ion channels.
Cloning novel channel genes
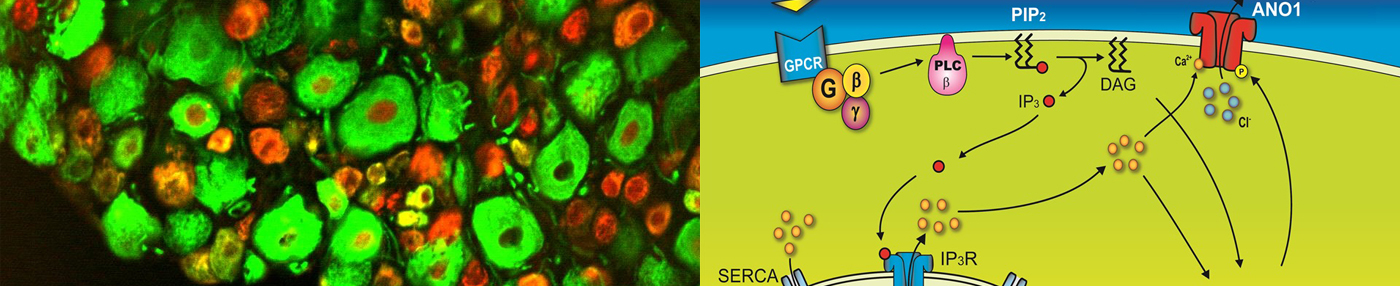
Channels are proteins present in the plasma membranes of all types of cells, including bacteria. Channels are activated by numerous stimuli such as electric voltage, mechanical agitation, heat, Ca2+ , and ligands. When activated, they adjust membrane potentials as well as intracellular Ca2+ concentration, thereby mediating numerous physiological functions. As a result, dysfunctions in these channels can result in numerous chronic diseases. The main focus of our laboratory is to clone novel channels using cutting-edge technologies such as bioinformatics.
Studying the biophysical properties of ion channels
Channels have their own unique properties: ion selectivity, activation mechanisms, pharmacology, modulation by cellular signals, etc. Using patch-clamp techniques, the ion channels are studied from cell lines, primary cultured cells or tissues in order to identify and characterize the channels’ biophysical properties.
Defining the functions of ion channels after ablating their genes.
By using various techniques for generating knock-out or knock-in mice, our laboratory seeks to define the physiological and pathological functions of channels. Anoctamins and mechanosensitive channels are our primary targets of study. And tissue-specific gene ablation in mice is preferred using the Cre-lox system.
Developing new drug candidates with channel modulators
Because channels mediate numerous key functions, defects can lead to chronic—often fatal—diseases. An example of this is cystic fibrosis, which results from loss-of-function mutations in a CFTR chloride channel. Therefore, chemicals that modulate channel activity are strong candidates for treating chronic diseases, which is why our center is diligently working to seek them out. Our center’s high throughput screening system will further aid us in finding new channel modulators from chemical libraries.